

The traditional methods for studying protein folding are minutely discussed. Protein folding is a thermodynamic process to create a 3D structure via minimum energy conformation based on entropy. The prognosis of protein 3D structure from the amino acid sequence has several applications in biological processes such as drug design, discovery of protein function, and interpretation of mutations in structural genomics. Because the experimental biologist suffered from the limited availability of 3D protein structure, protein structure prediction is effectively used to define 3D protein structure that supports more genetic information. Therefore, deep learning algorithms are applied to handling huge datasets for computational protein design by predicting the probability of 20 amino acids in a protein. Analysis of protein behavior can be difficult due to next-generation sequencing (NGS) technology, time-consuming, and low accuracy, especially for non-homologous protein sequences. According to the polypeptide structure, proteins are categorized into four classes: primary, secondary, tertiary, and quaternary. There are some factors that lead to mutations in the protein shape and lack of protein function, including temperature variations, pH, and chemical reactions. Therefore, there are various types of proteins according to their benefits and applications.
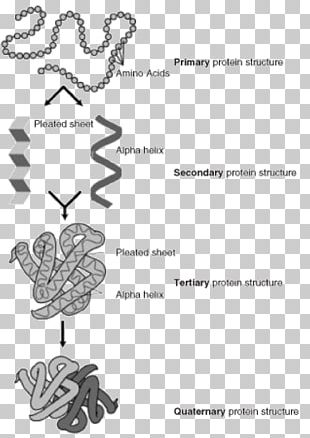
Proteins are diverse in shape and molecular weight and are relevant to their function and chemical bonds. Lastly, future work will focus mainly on large protein datasets without overfitting and expand the amount of extracted regulations for PSSP.
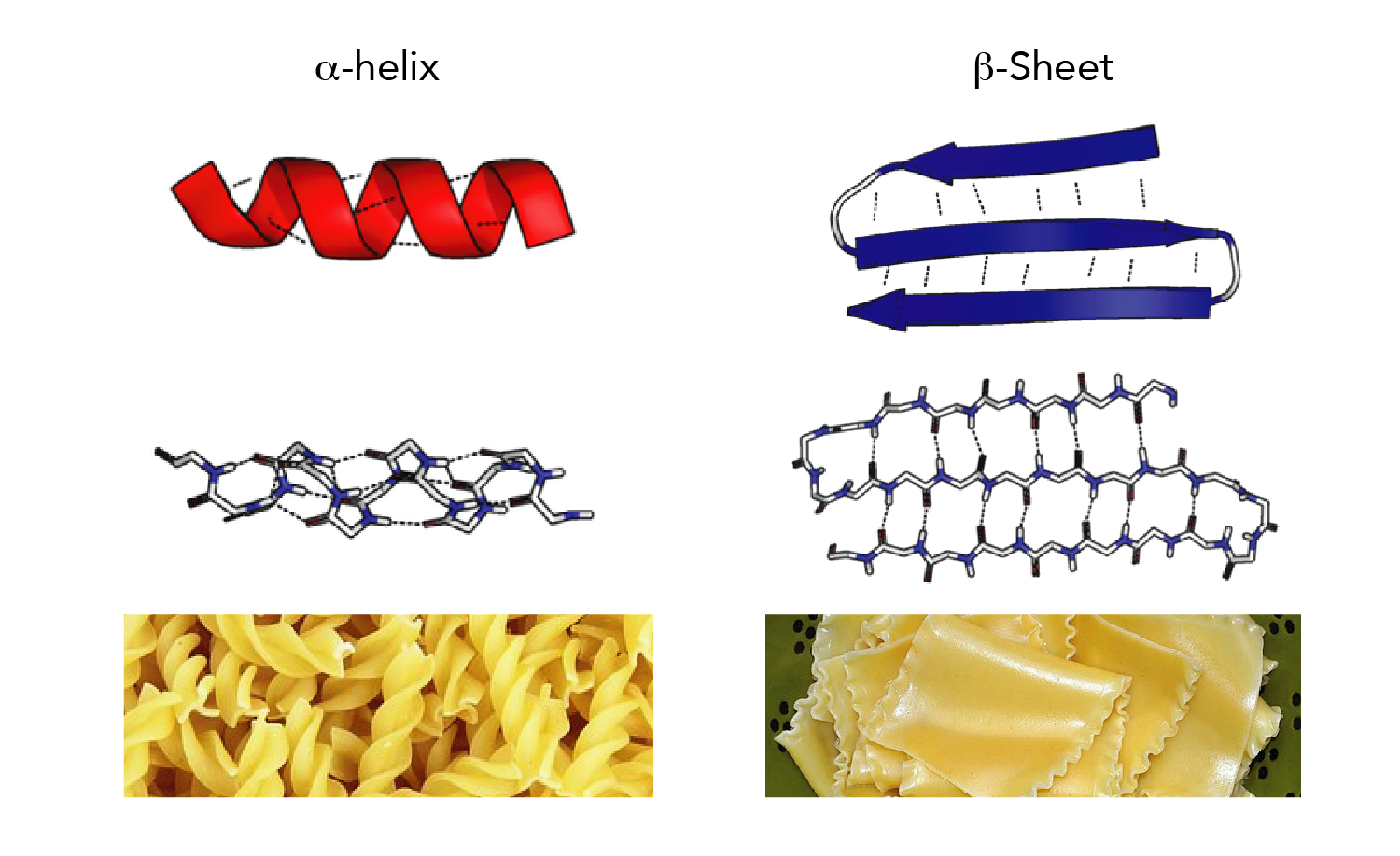
The proposed rules can be very important in managing wet laboratory experiments intended at determining protein secondary structure. The suggested algorithm achieved 85% accuracy for the E|~E classifier. The results confirmed that the existence of a particular amino acid in a protein sequence increases the stability for the forecast of protein secondary structure. Several of the proposed principles have compelling and relevant biological clarification. Furthermore, the number of produced rules was fairly small, and they show a greater degree of comprehensibility compared to other rules. A decision tree is applied for the SVM outcome to obtain the relevant guidelines possible for PSSP. In this paper, the support vector machine (SVM) model and decision tree are presented on the RS126 dataset to address the problem of PSSP. Extracting protein structure from the laboratory has insufficient information for PSSP that is used in bioinformatics studies. The computational biology approach has advanced exponentially in protein secondary structure prediction (PSSP), which is vital for the pharmaceutical industry.


 0 kommentar(er)
0 kommentar(er)
